With researchers touting recent success in sequencing the human genome’s remaining gaps, an emerging frontier is proteomics: identifying and studying an entire set of expressed proteins in the human body and other organisms. Collectively, these sets are called proteomes, and unlike genomes, proteomes alter over time and depict current health conditions—not conditions at risk of occurring (Video).
“The complexity in proteomics makes it a challenge,” Aleksandra Nita-Lazar, PhD, cochair of the National Institutes of Health’s Proteomics Scientific Interest Group, said in an interview with JAMA. “That's why it takes so long to get the complete proteomes for every organism, because proteins change a lot depending on conditions.”

Fig 1. Article title
In the meantime, the potential of proteomics in medicine is unfolding, as certain protein and proteome profiles are linked to disease. A recent study in PLOS Digital Health explored proteomic predictors of surviving COVID-19 in patients requiring intensive care. Studies in Scientific Reports as well as in eBioMedicine examined how proteome analysis of plasma may determine severity of COVID-19 cases.
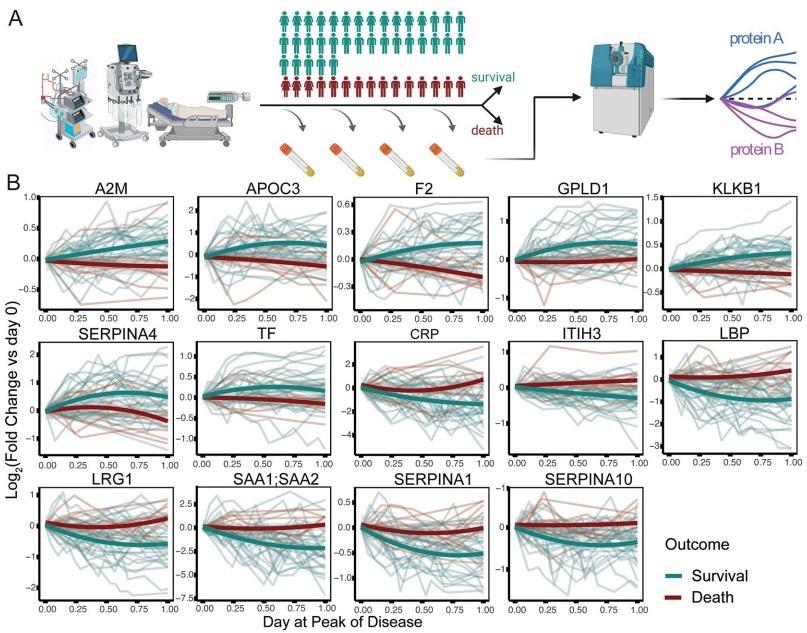
Fig 2. Prediction of survival or death in critically ill patients, from the first sampling time point at intensive care treatment level (WHO grade 7).
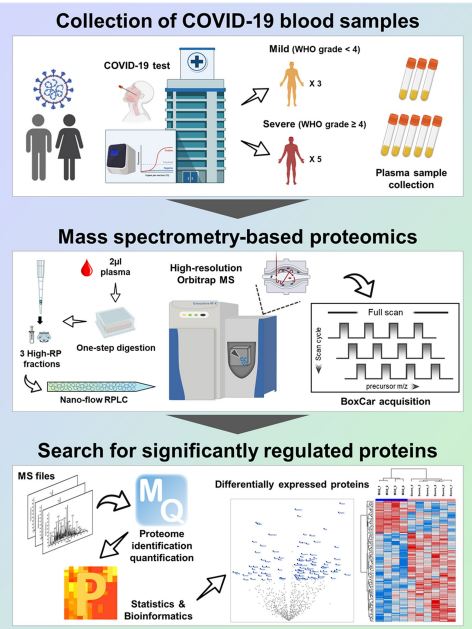
Fig 3. Overall scheme of in-depth plasma proteome profiling
Researchers have developed drafts of what’s known as the human proteome: the entire set of proteins expressed in the body. That human proteome is composed of subproteomes because each cell type has its own unique proteome. Then there are proteoforms, which make up a proteome, and these proteoforms are the protein variants—or forms—produced by a genome.
One gene can create an assortment of proteins with subtle differences. “For example, a human gene expressed in the liver, or the brain, or the kidney, or the hematopoietic system in your blood and bone marrow, these create all different proteoforms that present the unmapped frontier of the human proteome,” Neil Kelleher, PhD, director of Northwestern Proteomics and the Chemistry of Life Processes Institute at Northwestern University, said in an interview with JAMA.
Now, researchers aim to produce a reference set of proteoforms in the human body and hope to finish mapping the entire human proteome.
The Beginning
Two-dimensional (2D) gel electrophoresis—a technique used to separate, fractionate, and analyze proteins extracted from biological samples—helped make proteomics possible. Some of the first studies to encompass proteomics were published in 1975. Using the 2D gel technique, researchers mapped mouse, guinea pig, and Escherichia coli proteins—and separated the proteins by charge as well as size.
The field of proteins—and genomics—further progressed during the 1990s and early 2000s, as the first microbial genomes were sequenced and momentum grew for the Human Genome Project. Protein analysis relied less on 2D gels by the turn of the millennium, and advances in mass spectrometry allowed for analyzing ionized proteins in their gas phase. Researchers could now fragment proteins and identify them with more confidence. This also permitted work with complex protein mixtures rather than a single protein. However, a term to describe the study of proteins hadn’t yet worked its way into the lexicon.
Defining Moment
Although initial studies that could be considered proteomics were published nearly 50 years ago, a team of researchers including Marc Wilkins, DSc, PhD—currently a professor of systems biology at The University of New South Wales (UNSW) in Sydney, Australia—didn’t coin the term proteome until 1994. Then a doctoral student attending a 2D electrophoresis conference in Italy, Wilkins presented a coauthored paper on protein identification techniques. He also discussed new terminology.
“Whilst you could describe all the genes in the genome, there wasn't a way to do the same thing for proteins,” Wilkins recalled in an interview with JAMA. His solution: “It seemed obvious to combine proteinwith -ome, as it had been used in genome and biome.” Thus, the word proteome.
In 1995, Wilkins’ first journal article mentioning proteome was published in Electrophoresis. “'Proteome' refers to the total protein complement of a genome,” he and his colleagues wrote. Another article coauthored by Wilkins in Biotechnology and Genetic Engineering Reviews went further to define the term: “As an extrapolation of the concept of the 'genome project', a 'proteome project' is research which seeks to identify and characterise the proteins present in a cell or tissue and define their patterns of expression.”
Fast forward almost 2 decades: Kelleher and his colleague Lloyd Smith, PhD, a chemistry professor at the University of Wisconsin–Madison, propose the word proteoform “to designate all of the different molecular forms in which the protein product of a single gene can be found, including changes due to genetic variations, alternatively spliced RNA transcripts and post-translational modifications.” Justification for using the term was published in a 2013 issue of Nature Methods.

Fig 4. Dr.Marc Wilkins
What Counts
Various projects strive to complete the human proteome, each with distinct approaches. One of these endeavors is the Human Proteome Organization’s (HUPO) Human Proteome Project (HPP). As of March 2022, the HPP has discovered 93.2% of the human proteome—with 18 407 proteins identified—and estimates that the human genome encodes 19 750 proteins.
“The good news is that we can kind of predict the number of proteins that should be in the human proteome from the genome,” said Wilkins, a council member of HUPO who isn’t involved formally with the HPP.
Counting proteoforms is a larger endeavor because there are many more proteoforms than proteins. Thus far, the Human Proteoform Project has identified 61 770 proteoforms as of August 2022. But that’s not even close to the total number of proteoforms. “[B]asically, what we're talking about is defining something on the order of 50 [million] to 100 million unique proteoforms,” said Kelleher, who spearheads the Human Proteoform Project and is president of the board of directors for the Consortium for Top-Down Proteomics.
Going Forward
Some efforts offer protein data as they become available. The Human Protein Atlas is an open access database of categorized proteins, many based on antibodies, and includes images. The platforms UniProt and neXtProt also publish information about proteins while keeping track of which proteins still need additional research, such as those with unknown functions.
The open-sourced, artificial-intelligence system known as AlphaFold has predicted the shapes of most proteins thought to exist on Earth: more than 200 million. When researchers input a protein sequence, AlphaFold spits out a predicted structure. It’s especially important because a protein’s shape can influence its function.
“AlphaFold has been very helpful…we’re using it, too,” said Nita-Lazar, who’s also senior investigator of the National Institute of Allergy and Infectious Diseases’ National Cellular Networks Proteomic Unit and is on the executive committee of HUPO’s Biology & Disease-Driven Human Proteome Project. “It’s very interactive and collaborative.”
An open access database of structures is available online. Still, these predicted shapes remain just that: predictions. But from what researchers do know, there’s potential for applying proteomics to other fields, including medicine. For example, a proteoform-based assay can determine the species of infectious bacteria in patients.
Proteome analysis has been used for studying other illnesses as well. Misfolded proteins can lead to protein diseases known as proteinopathies, which include Alzheimer disease and Parkinson disease. Several studies have examined proteomic changes in the brains of patients with Alzheimer disease.
Another area using proteomics is cancer research: In 2011, the National Cancer Institute established the Clinical Proteomic Tumor Analysis Consortium to better understand cancer through proteogenomics—a method integrating both proteomics and genomics. The University of Texas MD Anderson Cancer Center also has a research platform for cancer proteomics, in hopes that it will advance its Moon Shots Program aimed at increasing cancer survival.
“Everyone in proteomics stands on each other's shoulders,” Wilkins said. “One of the things that has been fantastic to watch is the way that so many people with different approaches have come together to try and make this all happen.”

With researchers touting recent success in sequencing the human genome’s remaining gaps, an emerging frontier is proteomics: identifying and studying an entire set of expressed proteins in the human body and other organisms. Collectively, these sets are called proteomes, and unlike genomes, proteomes alter over time and depict current health conditions—not conditions at risk of occurring (Video).
“The complexity in proteomics makes it a challenge,” Aleksandra Nita-Lazar, PhD, cochair of the National Institutes of Health’s Proteomics Scientific Interest Group, said in an interview with JAMA. “That's why it takes so long to get the complete proteomes for every organism, because proteins change a lot depending on conditions.”

Fig 1. Article title
In the meantime, the potential of proteomics in medicine is unfolding, as certain protein and proteome profiles are linked to disease. A recent study in PLOS Digital Health explored proteomic predictors of surviving COVID-19 in patients requiring intensive care. Studies in Scientific Reports as well as in eBioMedicine examined how proteome analysis of plasma may determine severity of COVID-19 cases.

Fig 2. Prediction of survival or death in critically ill patients, from the first sampling time point at intensive care treatment level (WHO grade 7).

Fig 3. Overall scheme of in-depth plasma proteome profiling
Researchers have developed drafts of what’s known as the human proteome: the entire set of proteins expressed in the body. That human proteome is composed of subproteomes because each cell type has its own unique proteome. Then there are proteoforms, which make up a proteome, and these proteoforms are the protein variants—or forms—produced by a genome.
One gene can create an assortment of proteins with subtle differences. “For example, a human gene expressed in the liver, or the brain, or the kidney, or the hematopoietic system in your blood and bone marrow, these create all different proteoforms that present the unmapped frontier of the human proteome,” Neil Kelleher, PhD, director of Northwestern Proteomics and the Chemistry of Life Processes Institute at Northwestern University, said in an interview with JAMA.
Now, researchers aim to produce a reference set of proteoforms in the human body and hope to finish mapping the entire human proteome.
The Beginning
Two-dimensional (2D) gel electrophoresis—a technique used to separate, fractionate, and analyze proteins extracted from biological samples—helped make proteomics possible. Some of the first studies to encompass proteomics were published in 1975. Using the 2D gel technique, researchers mapped mouse, guinea pig, and Escherichia coli proteins—and separated the proteins by charge as well as size.
The field of proteins—and genomics—further progressed during the 1990s and early 2000s, as the first microbial genomes were sequenced and momentum grew for the Human Genome Project. Protein analysis relied less on 2D gels by the turn of the millennium, and advances in mass spectrometry allowed for analyzing ionized proteins in their gas phase. Researchers could now fragment proteins and identify them with more confidence. This also permitted work with complex protein mixtures rather than a single protein. However, a term to describe the study of proteins hadn’t yet worked its way into the lexicon.
Defining Moment
Although initial studies that could be considered proteomics were published nearly 50 years ago, a team of researchers including Marc Wilkins, DSc, PhD—currently a professor of systems biology at The University of New South Wales (UNSW) in Sydney, Australia—didn’t coin the term proteome until 1994. Then a doctoral student attending a 2D electrophoresis conference in Italy, Wilkins presented a coauthored paper on protein identification techniques. He also discussed new terminology.
“Whilst you could describe all the genes in the genome, there wasn't a way to do the same thing for proteins,” Wilkins recalled in an interview with JAMA. His solution: “It seemed obvious to combine proteinwith -ome, as it had been used in genome and biome.” Thus, the word proteome.
In 1995, Wilkins’ first journal article mentioning proteome was published in Electrophoresis. “'Proteome' refers to the total protein complement of a genome,” he and his colleagues wrote. Another article coauthored by Wilkins in Biotechnology and Genetic Engineering Reviews went further to define the term: “As an extrapolation of the concept of the 'genome project', a 'proteome project' is research which seeks to identify and characterise the proteins present in a cell or tissue and define their patterns of expression.”
Fast forward almost 2 decades: Kelleher and his colleague Lloyd Smith, PhD, a chemistry professor at the University of Wisconsin–Madison, propose the word proteoform “to designate all of the different molecular forms in which the protein product of a single gene can be found, including changes due to genetic variations, alternatively spliced RNA transcripts and post-translational modifications.” Justification for using the term was published in a 2013 issue of Nature Methods.

Fig4. Dr. Marc Wilkins
What Counts
Various projects strive to complete the human proteome, each with distinct approaches. One of these endeavors is the Human Proteome Organization’s (HUPO) Human Proteome Project (HPP). As of March 2022, the HPP has discovered 93.2% of the human proteome—with 18 407 proteins identified—and estimates that the human genome encodes 19 750 proteins.
“The good news is that we can kind of predict the number of proteins that should be in the human proteome from the genome,” said Wilkins, a council member of HUPO who isn’t involved formally with the HPP.
Counting proteoforms is a larger endeavor because there are many more proteoforms than proteins. Thus far, the Human Proteoform Project has identified 61 770 proteoforms as of August 2022. But that’s not even close to the total number of proteoforms. “[B]asically, what we're talking about is defining something on the order of 50 [million] to 100 million unique proteoforms,” said Kelleher, who spearheads the Human Proteoform Project and is president of the board of directors for the Consortium for Top-Down Proteomics.
Going Forward
Some efforts offer protein data as they become available. The Human Protein Atlas is an open access database of categorized proteins, many based on antibodies, and includes images. The platforms UniProt and neXtProt also publish information about proteins while keeping track of which proteins still need additional research, such as those with unknown functions.
The open-sourced, artificial-intelligence system known as AlphaFold has predicted the shapes of most proteins thought to exist on Earth: more than 200 million. When researchers input a protein sequence, AlphaFold spits out a predicted structure. It’s especially important because a protein’s shape can influence its function.
“AlphaFold has been very helpful…we’re using it, too,” said Nita-Lazar, who’s also senior investigator of the National Institute of Allergy and Infectious Diseases’ National Cellular Networks Proteomic Unit and is on the executive committee of HUPO’s Biology & Disease-Driven Human Proteome Project. “It’s very interactive and collaborative.”
An open access database of structures is available online. Still, these predicted shapes remain just that: predictions. But from what researchers do know, there’s potential for applying proteomics to other fields, including medicine. For example, a proteoform-based assay can determine the species of infectious bacteria in patients.
Proteome analysis has been used for studying other illnesses as well. Misfolded proteins can lead to protein diseases known as proteinopathies, which include Alzheimer disease and Parkinson disease. Several studies have examined proteomic changes in the brains of patients with Alzheimer disease.
Another area using proteomics is cancer research: In 2011, the National Cancer Institute established the Clinical Proteomic Tumor Analysis Consortium to better understand cancer through proteogenomics—a method integrating both proteomics and genomics. The University of Texas MD Anderson Cancer Center also has a research platform for cancer proteomics, in hopes that it will advance its Moon Shots Program aimed at increasing cancer survival.
“Everyone in proteomics stands on each other's shoulders,” Wilkins said. “One of the things that has been fantastic to watch is the way that so many people with different approaches have come together to try and make this all happen.”