DIA 蛋白质组学
DIA(Data-Independent Acquisition,数据非依赖性采集)是一种无歧视性和无随机性的蛋白质组分析技术, 将质谱全扫描范围分为若干个窗口,然后对每个窗口中的所有离子进行检测及碎裂,从而无遗漏无差异地获得样本中所有离子的信息,降低样本检测的缺失值,同时提高定量准确性和重复性,实现大样本队列中高稳定,高精准的蛋白质组定量分析。
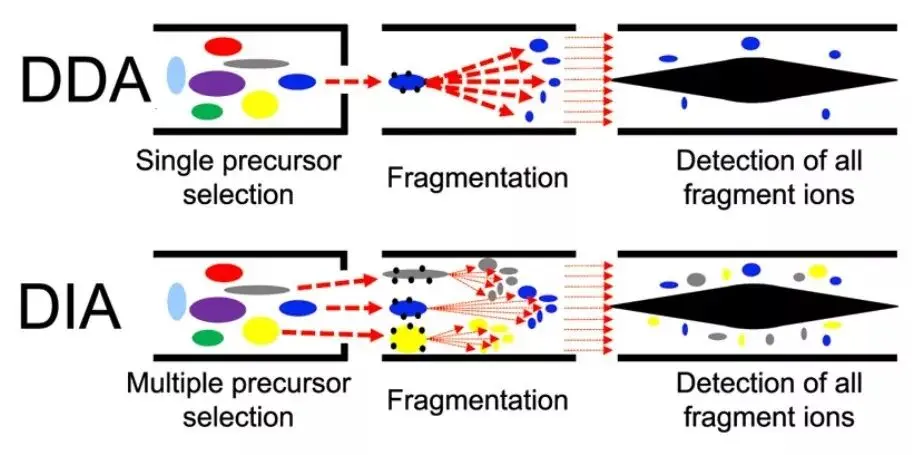
图1.1 DDA和DIA原理介绍(Hu A, Noble WS, Wolf-Yadlin A. Technical advances in proteomics: new developments in data-independent acquisition. F1000Res. 2016 Mar 31;5:F1000 Faculty Rev-419.)
4D-DIA 蛋白质组学
4D-DIA蛋白质组学是基于timsTOF Pro离子淌度平台的DIA技术,通过数据非依赖采集-同步累积连续碎裂(diaPASEF)扫描模式进行差异定量蛋白质组学分析。该技术核心是在传统3D分离(保留时间、质荷比、离子强度)基础上,增加肽段的碰撞截面积(CCS),实现第4维度(离子淌度)的分离检测。
将DIA技术与基于timsTOF Pro上的diaPASEF技术相结合,具有高扫描速度和高灵敏度优势的同时,外加第四维离子淌度分离,使DIA数据在采集时不牺牲窗口循环速度的同时,降低谱图复杂度和提高离子利用率,实现了蛋白质组学在覆盖深度、灵敏度、通量方面的全面性提升。
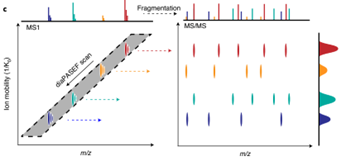
图1.2 4D-DIA原理介绍,离子淌度增加分离维度(Meier F, Brunner AD, Frank M, et al. diaPASEF: parallel accumulation-serial fragmentation combined with data-independent acquisition. Nat Methods. 2020 Dec;17(12):1229-1236.)
PulseDIA 蛋白质组学
PulseDIA脉冲式非依赖采集技术是西湖欧米专利技术,在传统DIA技术的基础上,将一个窗口根据质荷比分割成更小范围的多个窗口,并且将这些窗口以脉冲的形式均匀分配至不同的质谱方法进行采集,在相同时间下较DIA能提升蛋白鉴定深度10%-30%。
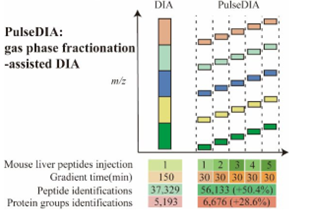
图1.3 PulseDIA原理介绍(Cai X, Ge W, Yi X, et al. PulseDIA: Data-Independent Acquisition Mass Spectrometry Using Multi-Injection Pulsed Gas-Phase Fractionation. J Proteome Res. 2021 Jan 1;20(1):279-288. )
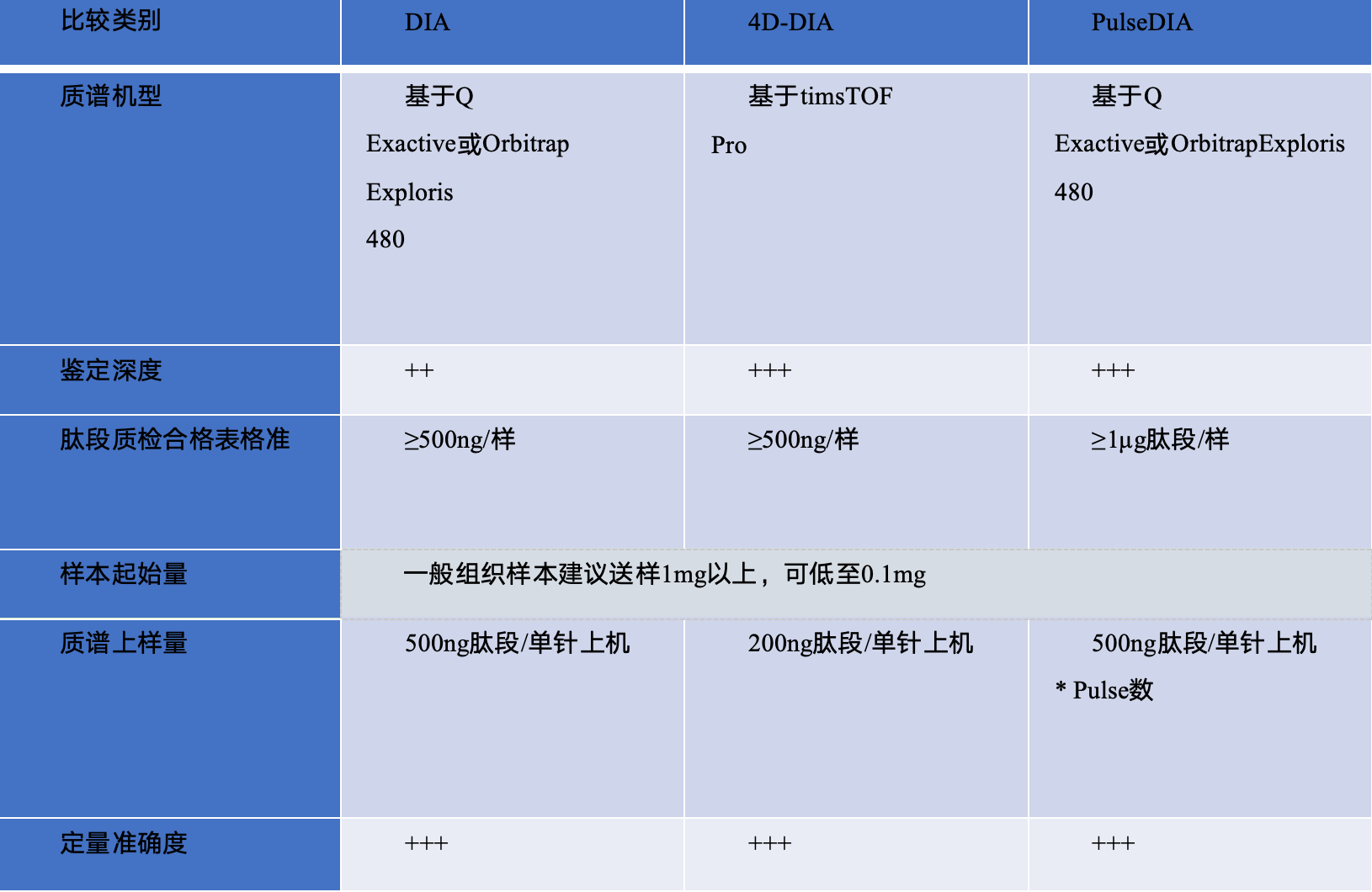
合作项目案例
Cancer Communications | DIA 技术解析结直肠癌肿瘤组织 FFPE 样本蛋白组特征,揭示 PLOD2 是结直肠癌潜在治疗新靶点
该研究利用 PCT 微量样本前处理技术和 DIA 技术结合,在结直肠癌患者的 170 个 FFPE 组织样本中量化了 6359 种蛋白质,技术重复之间的相关系数为 0.953。该研究在不同临床阶段结直肠癌患者肿瘤组织样本和正常结肠组织样本中分析出表达水平显著差异的 928 种蛋白质,并进一步通过靶向表达水平验证和细胞水平的功能验证明确了 PLOD2 为潜在的 CRC 治疗靶点。
参考文献
2. Röst et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nature Methods. 2016.13(9):741-748
https://www.nature.com/articles/nmeth.3959
3.Röst, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nature Biotechnology. 2014.32:219-223
https://www.nature.com/articles/nbt.2841
4.Guo, et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nature Medicine. 2015.21(4):407–413.
https://www.nature.com/articles/nm.3807
5. Searle et al. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nature Communications. 2018. 9(1): 1-12
https://www.nature.com/articles/s41467-018-07454-w
6.Xu, et al. In-depth Serum Proteomics Reveals Biomarkers of Psoriasis Severity and Response to Traditional Chinese Medicine. Theranostics. 2019.9(9): 2475-2488.
https://www.thno.org/v09p2475.htm
7.Shao, et al. Comparative analysis of mRNA and protein degradation in prostate tissues indicates high stability of proteins. Nature Communications. 2019. 10(1):2524.
https://www.nature.com/articles/s41467-019-10513-5
8.Zhu, et al. High-throughput Proteomic analysis of FFPE tissue samples facilitates tumor stratification. Molecular Oncology. 2019 Sep;13(11): 2305-2328.
https://febs.onlinelibrary.wiley.com/doi/10.1002/1878-0261.12570
9.Zhang, et al. Data-Independent Acquisition Mass Spectrometry-Based Proteomics and Software Tools: A Glimpse in 2020. Proteomics. 2020.20(17-18): e1900276.
https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/pmic.201900276
10.Demichev, et al. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nature Methods. 2020.17:41-44
https://www.nature.com/articles/s41592-019-0638-x
11.Cai, et al. PulseDIA: Data-Independent Acquisition Mass Spectrometry Using Multi-Injection Pulsed Gas-Phase Fractionation. Journal of Proteome Research. 2021.20(1):279-288.
https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00381
12.Ge, et al. Computational Optimization of Spectral Library Size Improves DIA-MS Proteome Coverage and Applications to 15 Tumors. Journal of Proteome Research. 2021
https://pubs.acs.org/doi/full/10.1021/acs.jproteome.1c00640
13.Liu, et al. DIA-based Proteomics Identifies IDH2 as a Targetable Regulator of Acquired Drug Resistance in Chronic Myeloid Leukemia. Mol Cell Prot. available at bioRxiv, 2021.
https://www.mcponline.org/article/S1535-9476(21)00159-6/fulltext#secsectitle0030
14.Shao, et al. Proteomics profiling of colorectal cancer progression identifies PLOD2 as a potential therapeutic target. Cancer Commun. 2021.
https://onlinelibrary.wiley.com/doi/10.1002/cac2.12240
15.Zhu, et al. Snapshot: Clinical proteomics. Cell. 2021.184(18): 4840-4840.